WEEK FIVE
- MONDAY 24/7/17 -
This morning, I carried out the repeat FACS experiment with the A549 cells. None of the antibodies recognised endogenous Tspan14 on A549 cells. 9D5 produced a more positive shift than in the previous experiment, however it is probably an insignificant result as the shift is not as large as the one with 5D4.
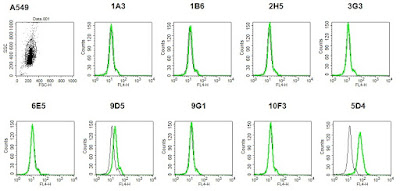 |
| Experiment: Do the Tspan14 mAbs recognise endogenous Tspan14 on A549 cells? |
In the afternoon, Connie showed me how to seed the HEK cells for immunostaining. I was very surprised I managed to seed all 20 wells without breaking a single coverslip! :)
- TUESDAY 25/7/17 -
 |
| Aspirating PBS from cells |
 |
| The cells were fixed with formalin for 15 mins. |
 |
I checked on the HEK cells to see if they were growing. I seeded 5 x 6cm plates and also had enough cells to do a 1:5 split to keep the cells growing.
In the afternoon, I went to the med school to do some imaging with Connie using the confocal microscope.
ADVANTAGES OF CONFOCAL MICROSCOPY:
Today, I transfected the HEK cells with the following constructs ready for the big FACS experiment tomorrow:
In the afternoon, I went to the med school to do some imaging with Connie using the confocal microscope.
ADVANTAGES OF CONFOCAL MICROSCOPY:
- Can control depth of field
- The pinhole only allows in focus light to pass through into the detector and blocks light from above and below the focal plane.
- Can optically slice through the sample and create a 3D reconstrution
- Can zoom into sample without losing resolution
 |
| Leica Confocal Microscop |
It took a very long time for me to get used to using the microscope and I had a lot of help from Connie with focusing and finding the cells. There weren't many cells on the hTspan14 coverslips, as most of them were washed off, which made them almost impossible to find!
SUMMARY OF RESULTS
Only 10F3 gave a strong signal with the endogenous Tspan14, which was very strange as none of the antibodies had been recognising Tspan14 using FACS on wild type HEK cells. However, the signal wasn't consistent across the whole coverslip and was only bright with a few cells. There was no signal for the other antibodies with the wild type cells, as expected. There was some signal with the overexpressed cells, however it wasn't as bright as expected.
We will do a repeat next week with the WT cells at a higher intensity to see if the antibodies are able to recognise endogenous Tspan14. I also need to make a new coverslip for overexpressed cells with 9D5, as I managed to snap and lose half of the coverslip during the washing step😂
We will do a repeat next week with the WT cells at a higher intensity to see if the antibodies are able to recognise endogenous Tspan14. I also need to make a new coverslip for overexpressed cells with 9D5, as I managed to snap and lose half of the coverslip during the washing step😂
- THURSDAY 27/7/17 -
 |
| Waiting for 10mins to allow DNA/PEI complexes to form. |
1. pEF6 - His
2. pEF6 FLAG hTspan14
3. pEF6 FLAG hCD9
4. pEF6 FLAG hCD9/Tspan14 EC2
5. pEF6 FLAG hCD9/Tspan14 var
- FRIDAY 28/7/17 -
Today I did the big FACS experiment to see if the Tspan14 mAbs recognise the main extracellular region (EC2) or specifically the variable region (var) of human Tspan14.
And as always, I ended up with very strange data.
 |
| As expected, none of the antibodies recognised endogenous Tspan14 |
 |
| None of the antibodies recognised CD9 |
 |
| 2H5 and 9D5 didn't recognise overexpressed Tspan14 |
 |
| None of the antibodies recognised the variable region of Tspan14 |
 |
| Some of the antibodies recognised the extracellular region of Tspan14 |



Comments
Post a Comment